I haven’t blogged since a long long time, on this opportunity I want to show a very simple way to obtain the nearest centroid.
Let’s imagine there is a group of markets in a town and they need to obtain their supply at the nearest supermarket.
With the help of a GIS Software, any facility has their geographical position and consecutively we can obtain a distance matrix. We assume that the supply only comes from only one supermarket. With this elements at hand we can make a code in R.
First of all to be a reproducible example we simulate a distance matrix. For this I can create and bring a matrix to a data frame. As we’ll on the code, I’m assuming that the random distances have a normal distribution.
As the output of a distance matrix of a GIS is tabular and it has not as a matrix , I have to convert the matrix to tabular data, for that I need to use the gather function found in the dplyr library.
Dist_mtx <- as.data.frame(matrix(abs(rnorm(100, mean = 1000, sd = 800)), nrow = 20))
names(Dist_mtx) <- seq(1:5)
Dist_mtx$O <- seq(10000, 10019, 1)
library(dplyr)
Dist_mtx <- gather(Dist_mtx, key = D, value = DIST, 1:5)
Dist_mtx <- Dist_mtx[order(Dist_mtx$O),]
After that there is the need to create a function with three parameters, we'll call her DistMin_Cent.
DistMin_Cent <- function(DF, Destination, Distance){
}
For this function we will need the libraries dplyr and tidyr because there is the need to transform the tabular data into a matrix form. This need of this transformation exist to extract the closest supermarket, this is with the spread function.
DF <- spread_(DF, Destination, Distance)
To extract the min distance we need the apply function, inside this function we need to set the argument MARGIN = 1, this is necessary to do the calculation per row.
DF$MIN <- apply(DF[, c(2:ncol(DF))], 1, FUN = min)
The following code helps to obtain to identify the name of the column which has the minimum distance.
c_col <- c(ncol(DF)-1)
DF$CName <- as.numeric(colnames(DF[, c(2:c_col)])[apply(DF[,
c(2:c_col)], 1, which.min)])
After that we select only the columns is useful for us.
All is summarized in a the new function to be capable of doing this with any data frame.
DistMin_Cent <- function(DF, Destination, Distance){
library(dplyr)
library(tidyr)
DF <- spread_(DF, Destination, Distance)
DF$MIN <- apply(DF[, c(2:ncol(DF))], 1, FUN = min)
c_col <- c(ncol(DF)-1)
DF$CName <- as.numeric(colnames(DF[, c(2:c_col)])[apply(DF[,
c(2:c_col)], 1, which.min)])
DF <- DF[, c(1, ncol(DF), c_col)]
rm(c_col)
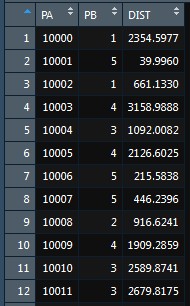
names(DF) <- c("PA", "PB", "DIST")
DF
}
DistMin_Cent(Dist_mtx, "D", "DIST")
And then we can see the results of the process.